🎯 | Motivation¶
- In this notebook, we will demonstrate the usage of the multi-backend capabilities of
KerasCoreandKerasNLPfor the MultipleChoice task. Our aim is to provide a comprehensive guide to building a MultipleChoice model using pre-trained NLP models from theKerasNLPlibrary. - While there are already numerous notebooks showcasing how to employ popular models like
Bert/DebertawithAutoModelForMultipleChoicefrom HuggingFace, these notebooks excel in quick training and inference. However, understanding the inner workings of the model might be challenging for those new to the HF library. Our notebook seeks to offer a deeper insight into constructing a MultipleChoice model using pre-trained NLP models from theKerasNLPlibrary. - Thanks to to
KerasCore, which enables seamless execution of this notebook onTensorFlow,Jax, andPyTorchplatforms with minimal adjustments required. - Moreover, this notebook accommodates both Single/Multi GPU and TPU training. As time progresses, larger datasets may become available, making TPUs invaluable for training substantial models on these extensive datasets.
📌 | Updates¶
v11-DebertaV3BaseEnglish modelv10- FixShuffleOptionaugmentation; answers need to be shuffled in same orderv09- TPU withjaxbacked but low lrv08- TPU withjaxbackend
🛠 | Install Libraries¶
!pip install -q keras-core --upgrade
!pip install -q keras-nlp --upgrade
!pip install -q wandb --upgrade
📚 | Import Libraries¶
import os
os.environ["KERAS_BACKEND"] = "jax" # or "tensorflow" or "torch"
os.environ["WANDB_SILENT"] = "true" # for wandb
import keras_nlp
import keras_core as keras
import keras_core.backend as K
import jax
import tensorflow as tf
# from tensorflow import keras
# import tensorflow.keras.backend as K
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib as mpl
cmap = mpl.cm.get_cmap('coolwarm')
Using JAX backend.
/opt/conda/lib/python3.10/site-packages/scipy/__init__.py:146: UserWarning: A NumPy version >=1.16.5 and <1.23.0 is required for this version of SciPy (detected version 1.23.5
warnings.warn(f"A NumPy version >={np_minversion} and <{np_maxversion}"
/opt/conda/lib/python3.10/site-packages/tensorflow_io/python/ops/__init__.py:98: UserWarning: unable to load libtensorflow_io_plugins.so: unable to open file: libtensorflow_io_plugins.so, from paths: ['/opt/conda/lib/python3.10/site-packages/tensorflow_io/python/ops/libtensorflow_io_plugins.so']
caused by: ['/opt/conda/lib/python3.10/site-packages/tensorflow_io/python/ops/libtensorflow_io_plugins.so: undefined symbol: _ZN3tsl6StatusC1EN10tensorflow5error4CodeESt17basic_string_viewIcSt11char_traitsIcEENS_14SourceLocationE']
warnings.warn(f"unable to load libtensorflow_io_plugins.so: {e}")
/opt/conda/lib/python3.10/site-packages/tensorflow_io/python/ops/__init__.py:104: UserWarning: file system plugins are not loaded: unable to open file: libtensorflow_io.so, from paths: ['/opt/conda/lib/python3.10/site-packages/tensorflow_io/python/ops/libtensorflow_io.so']
caused by: ['/opt/conda/lib/python3.10/site-packages/tensorflow_io/python/ops/libtensorflow_io.so: undefined symbol: _ZTVN10tensorflow13GcsFileSystemE']
warnings.warn(f"file system plugins are not loaded: {e}")
/tmp/ipykernel_23/4044669206.py:21: MatplotlibDeprecationWarning: The get_cmap function was deprecated in Matplotlib 3.7 and will be removed two minor releases later. Use ``matplotlib.colormaps[name]`` or ``matplotlib.colormaps.get_cmap(obj)`` instead.
cmap = mpl.cm.get_cmap('coolwarm')
Library Version¶
print("TensorFlow:", tf.__version__)
print("JAX:", jax.__version__)
print("Keras:", keras.__version__)
print("KerasNLP:", keras_nlp.__version__)
TensorFlow: 2.12.0 JAX: 0.4.13 Keras: 0.1.4 KerasNLP: 0.6.1
⚙️ | Configuration¶
class CFG:
verbose = 0 # Verbosity
wandb = True # Weights & Biases logging
competition = 'kaggle-llm-science-exam' # Competition name
_wandb_kernel = 'awsaf49' # WandB kernel
comment = 'DebertaV3-MaxSeq_200-ext_s-low_lr-aug_fix' # Comment description
preset = "deberta_v3_base_en" # Name of pretrained models
sequence_length = 200 # Input sequence length
device = 'TPU' # Device
seed = 42 # Random seed
num_folds = 5 # Total folds
selected_folds = [3, 4] # Folds to train on
epochs = 10 # Training epochs
batch_size = 2 # Batch size
drop_remainder = True # Drop incomplete batches
cache = True # Caches data after one iteration, use only with `TPU` to avoid OOM
augment = True # Augmentation (Shuffle Options)
scheduler = 'cosine' # Learning rate scheduler
external_data = True # External data flag
class_names = list("ABCDE") # Class names [A, B, C, D, E]
num_classes = len(class_names) # Number of classes
class_labels = list(range(num_classes)) # Class labels [0, 1, 2, 3, 4]
label2name = dict(zip(class_labels, class_names)) # Label to class name mapping
name2label = {v: k for k, v in label2name.items()} # Class name to label mapping
♻️ | Reproducibility¶
Sets value for random seed to produce similar result in each run.
keras.utils.set_random_seed(CFG.seed)
💾 | Hardware¶
Following codes automatically detects hardware (TPU or GPU).
def get_device():
"Detect and intializes GPU/TPU automatically"
try:
# Connect to TPU
tpu = tf.distribute.cluster_resolver.TPUClusterResolver.connect()
# Set TPU strategy
strategy = tf.distribute.TPUStrategy(tpu)
print(f'> Running on TPU', tpu.master(), end=' | ')
print('Num of TPUs: ', strategy.num_replicas_in_sync)
device=CFG.device
except:
# If TPU is not available, detect GPUs
gpus = tf.config.list_logical_devices('GPU')
ngpu = len(gpus)
# Check number of GPUs
if ngpu:
# Set GPU strategy
strategy = tf.distribute.MirroredStrategy(gpus) # single-GPU or multi-GPU
# Print GPU details
print("> Running on GPU", end=' | ')
print("Num of GPUs: ", ngpu)
device='GPU'
else:
# If no GPUs are available, use CPU
print("> Running on CPU")
strategy = tf.distribute.get_strategy()
device='CPU'
return strategy, device
# Initialize GPU/TPU/TPU-VM
strategy, CFG.device = get_device()
CFG.replicas = strategy.num_replicas_in_sync
> Running on GPU | Num of GPUs: 1
📁 | Dataset Path¶
BASE_PATH = '/kaggle/input/kaggle-llm-science-exam'
📖 | Meta Data¶
- train.csv - a set of 200 questions with the answer column. Each question consists of a
prompt(the question), 5 options labeledA,B,C,D, andE, and the correct answer labeledanswer(this holds the label of the most correct answer, as defined by the generating LLM). - test.csv - similar to train.csv except it doesn't have
answercolumn. It has ~4000 questions that may be different is subject matter. - sample_submission.csv - is the valid sample submission.
id: number id of the questionprediction: top 3 labels for your prediction. Once a correct label has been scored for an individual question in the test set, that label is no longer considered relevant for that question, and additional predictions of that label are skipped in the calculation
Train Data¶
df = pd.read_csv(f'{BASE_PATH}/train.csv') # Read CSV file into a DataFrame
df['label'] = df.answer.map(CFG.name2label) # Map answer labels using name-to-label mapping
# Display information about the train data
print("# Train Data: {:,}".format(len(df)))
print("# Sample:")
display(df.head(2))
# Show distribution of answers using a bar plot
plt.figure(figsize=(8, 4))
df.answer.value_counts().plot.bar(color=[cmap(0.0), cmap(0.25), cmap(0.65), cmap(0.9), cmap(1.0)])
plt.xlabel("Answer")
plt.ylabel("Count")
plt.title("Answer distribution for Train Data")
plt.show()
# Train Data: 200 # Sample:
| id | prompt | A | B | C | D | E | answer | label | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | Which of the following statements accurately d... | MOND is a theory that reduces the observed mis... | MOND is a theory that increases the discrepanc... | MOND is a theory that explains the missing bar... | MOND is a theory that reduces the discrepancy ... | MOND is a theory that eliminates the observed ... | D | 3 |
| 1 | 1 | Which of the following is an accurate definiti... | Dynamic scaling refers to the evolution of sel... | Dynamic scaling refers to the non-evolution of... | Dynamic scaling refers to the evolution of sel... | Dynamic scaling refers to the non-evolution of... | Dynamic scaling refers to the evolution of sel... | A | 0 |
External Datasets¶
We'll be utilizing the following external datasets:
# Concatenate multiple external datasets into one DataFrame
ext_df = pd.concat([
pd.read_csv('/kaggle/input/additional-train-data-for-llm-science-exam/extra_train_set.csv'),
# pd.read_csv('/kaggle/input/additional-train-data-for-llm-science-exam/6000_train_examples.csv'),
# pd.read_csv('/kaggle/input/llm-science-3k-data/test.csv'),
# pd.read_csv('/kaggle/input/wikipedia-stem-1k/stem_1k_v1.csv')
])
# ext_df.drop(columns=["id"], inplace=True) # Drop 'id' column
ext_df.reset_index(drop=True, inplace=True) # Reset index
ext_df["id"] = ext_df.index.tolist() # Add 'id' column as index
ext_df['label'] = ext_df.answer.map(CFG.name2label) # Map answer labels using name-to-label mapping
# Display information about the external data
print("# External Data: {:,}".format(len(ext_df)))
print("# Sample:")
ext_df.head(2)
# Show distribution of answers using a bar plot
plt.figure(figsize=(8, 4))
ext_df.answer.value_counts().plot.bar(color=[cmap(0.0), cmap(0.25), cmap(0.65), cmap(0.9), cmap(1.0)])
plt.xlabel("Answer")
plt.ylabel("Count")
plt.title("Answer distribution for External Data")
plt.show()
# External Data: 500 # Sample:
Contextualize Options¶
Our approach entails furnishing the model with question and answer pairs, as opposed to employing a single question for all five options. In practice, this signifies that for the five options, we will supply the model with the same set of five questions combined with each respective answer choice (e.g., (Q + A), (Q + B), and so on). This analogy draws parallels to the practice of revisiting a question multiple times during an exam to promote a deeper understanding of the problem at hand.
# Define a function to create options based on the prompt and choices
def make_options(row):
row['options'] = [f"{row.prompt}\n{row.A}", # Option A
f"{row.prompt}\n{row.B}", # Option B
f"{row.prompt}\n{row.C}", # Option C
f"{row.prompt}\n{row.D}", # Option D
f"{row.prompt}\n{row.E}"] # Option E
return row
df = df.apply(make_options, axis=1) # Apply the make_options function to each row in df
df.head(2) # Display the first 2 rows of df
ext_df = ext_df.apply(make_options, axis=1) # Apply the make_options function to each row in ext_df
ext_df.head(2) # Display the first 2 rows of ext_df
| prompt | C | E | D | B | A | answer | id | label | options | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | In relation to Eunice Fay McKenzie's career, w... | McKenzie gained recognition for her role as a ... | McKenzie's successful career in sound films co... | McKenzie's collaborations with director Blake ... | McKenzie is primarily remembered for her starr... | McKenzie showcased her singing talents in nume... | B | 0 | 1 | [In relation to Eunice Fay McKenzie's career, ... |
| 1 | How does Modified Newtonian Dynamics (MOND) im... | MOND is a theory that reduces the observed mis... | MOND's impact on the observed missing baryonic... | MOND is a theory that eliminates the observed ... | MOND explains the missing baryonic mass in gal... | MOND is a theory that increases the discrepanc... | E | 1 | 4 | [How does Modified Newtonian Dynamics (MOND) i... |
🔪 | Data Split¶
In the code snippet provided below, we will divide the existing train data into folds using a stratification of answer column.
It's worth noting that external data will not find usage in the validation phase. Instead, for every fold, the external data will only contribute to the training process. To elaborate, consider the scenario of fold0 training:
- Training data: fold1, fold2, fold3, external
- Validation data: fold0
from sklearn.model_selection import StratifiedKFold # Import package
skf = StratifiedKFold(n_splits=CFG.num_folds, shuffle=True, random_state=CFG.seed) # Initialize K-Fold
df = df.reset_index(drop=True) # Reset dataframe index
df["fold"] = -1 # New 'fold' column
# Assign folds using StratifiedKFold
for fold, (train_idx, val_idx) in enumerate(skf.split(df, df['answer'])):
df.loc[val_idx, 'fold'] = fold
# Display label distribution for each fold
df.groupby(["fold", "answer"]).size()
fold answer
0 A 7
B 10
C 9
D 8
E 6
1 A 7
B 10
C 9
D 8
E 6
2 A 7
B 9
C 9
D 8
E 7
3 A 8
B 9
C 9
D 7
E 7
4 A 8
B 10
C 8
D 7
E 7
dtype: int64 🍽️ | Preprocessing¶
What it does: The preprocessor takes input strings and transforms them into a dictionary (token_ids, padding_mask) containing preprocessed tensors. This process starts with tokenization, where input strings are converted into sequences of token IDs.
Why it's important: Initially, raw text data is complex and challenging for modeling due to its high dimensionality. By converting text into a compact set of tokens, such as transforming "The quick brown fox" into ["the", "qu", "##ick", "br", "##own", "fox"], we simplify the data. Many models rely on special tokens and additional tensors to understand input. These tokens help divide input and identify padding, among other tasks. Making all sequences the same length through padding boosts computational efficiency, making subsequent steps smoother.
Explore the following pages to access the available preprocessing and tokenizer layers in KerasNLP:
preprocessor = keras_nlp.models.DebertaV3Preprocessor.from_preset(
preset=CFG.preset, # Name of the model
sequence_length=CFG.sequence_length, # Max sequence length, will be padded if shorter
)
Downloading data from https://storage.googleapis.com/keras-nlp/models/deberta_v3_base_en/v1/vocab.spm 2464616/2464616 ━━━━━━━━━━━━━━━━━━━━ 1s 0us/step
Now, let's examine what the output shape of the preprocessing layer looks like. The output shape of the layer can be represented as $(num\_choices, sequence\_length)$.
outs = preprocessor(df.options.iloc[0]) # Process options for the first row
# Display the shape of each processed output
for k, v in outs.items():
print(k, ":", v.shape)
token_ids : (5, 200) padding_mask : (5, 200)
We'll use the preprocessing_fn function to transform each text option using the dataset.map(preprocessing_fn) method.
def preprocess_fn(text, label=None):
text = preprocessor(text) # Preprocess text
return (text, label) if label is not None else text # Return processed text and label if available
🌈 | Augmentation¶
OptionShuffle¶
In this notebook, we'll experiment with an interesting augmentation technique, OptionShuffle. Since we're providing the model with one option at a time, we can introduce a shuffle to the order of options. For instance, options [A, C, E, D, B] would be rearranged as [D, B, A, E, C]. This practice will help the model focus on the content of the options themselves, rather than being influenced by their positions.
def OptionShuffle(options, labels, prob=0.50, seed=None):
if tf.random.uniform([]) > prob: # Shuffle probability check
return options, labels
# Shuffle indices of options and labels in the same order
indices = tf.random.shuffle(tf.range(tf.shape(options)[0]), seed=seed)
# Shuffle options and labels
options = tf.gather(options, indices)
labels = tf.gather(labels, indices)
return options, labels
In the following function, we'll merge all augmentation functions to apply to the text. These augmentations will be applied to the data using the dataset.map(augment_fn) approach.
def augment_fn(text, label=None):
text, label = OptionShuffle(text, label, prob=0.5) # Apply OptionShuffle
return (text, label) if label is not None else text
🍚 | DataLoader¶
The code below sets up a robust data flow pipeline using tf.data.Dataset for data processing. Notable aspects of tf.data include its ability to simplify pipeline construction and represent components in sequences.
To learn more about tf.data, refer to this documentation.
def build_dataset(texts, labels=None, batch_size=32,
cache=False, drop_remainder=True,
augment=False, repeat=False, shuffle=1024):
AUTO = tf.data.AUTOTUNE # AUTOTUNE option
slices = (texts,) if labels is None else (texts, keras.utils.to_categorical(labels, num_classes=5)) # Create slices
ds = tf.data.Dataset.from_tensor_slices(slices) # Create dataset from slices
ds = ds.cache() if cache else ds # Cache dataset if enabled
if augment: # Apply augmentation if enabled
ds = ds.map(augment_fn, num_parallel_calls=AUTO)
ds = ds.map(preprocess_fn, num_parallel_calls=AUTO) # Map preprocessing function
ds = ds.repeat() if repeat else ds # Repeat dataset if enabled
opt = tf.data.Options() # Create dataset options
if shuffle:
ds = ds.shuffle(shuffle, seed=CFG.seed) # Shuffle dataset if enabled
opt.experimental_deterministic = False
ds = ds.with_options(opt) # Set dataset options
ds = ds.batch(batch_size, drop_remainder=drop_remainder) # Batch dataset
ds = ds.prefetch(AUTO) # Prefetch next batch
return ds # Return the built dataset
Fetch Train/Valid Dataset¶
The function below generates the training and validation datasets for a given fold.
def get_datasets(fold):
train_df = df.query("fold!=@fold") # Get training fold data
if CFG.external_data:
train_df = pd.concat([train_df, ext_df], axis=0) # Add external data texts
train_df = train_df.reset_index(drop=True)
train_texts = train_df.options.tolist() # Extract training texts
train_labels = train_df.label.tolist() # Extract training labels
# Build training dataset
train_ds = build_dataset(train_texts, train_labels,
batch_size=CFG.batch_size*CFG.replicas, cache=CFG.cache,
shuffle=True, drop_remainder=True, repeat=True, augment=CFG.augment)
valid_df = df.query("fold==@fold") # Get validation fold data
valid_texts = valid_df.options.tolist() # Extract validation texts
valid_labels = valid_df.label.tolist() # Extract validation labels
# Build validation dataset
valid_ds = build_dataset(valid_texts, valid_labels,
batch_size=min(CFG.batch_size*CFG.replicas, len(valid_df)), cache=CFG.cache,
shuffle=False, drop_remainder=True, repeat=False, augment=False)
return (train_ds, train_df), (valid_ds, valid_df) # Return datasets and dataframes
🪄 | Wandb¶
To monitor the training of my text-based model, I'll make use of Weights & Biases. Weights & Biases (W&B) is an MLOps platform that offers experiment tracking, dataset versioning, and model management functionalities, aiding in efficient model development.
import wandb # Import wandb library for experiment tracking
try:
from kaggle_secrets import UserSecretsClient # Import UserSecretsClient
user_secrets = UserSecretsClient() # Create secrets client instance
api_key = user_secrets.get_secret("WANDB") # Get API key from Kaggle secrets
wandb.login(key=api_key) # Login to wandb with the API key
anonymous = None # Set anonymous mode to None
except:
anonymous = 'must' # Set anonymous mode to 'must'
wandb.login(anonymous=anonymous, relogin=True) # Login to wandb anonymously and relogin if needed
Logger¶
The following code cell contains code to log data to WandB. It is noteworthy that the newly released callbacks offer more flexibility in terms of customization, and they are more compact compared to the classic WandbCallback, making it easier to use. Here's a brief introduction to them:
- WandbModelCheckpoint: This callback saves the model or weights using
tf.keras.callbacks.ModelCheckpoint. Hence, we can harness the power of the official TensorFlow callback to log eventf.keras.Modelsubclass model in TPU. - WandbMetricsLogger: This callback simply logs all the metrics and losses.
- WandbEvalCallback: This one is even more special. We can use it to log the model's prediction after a certain epoch/frequency. We can use it to save segmentation masks, bounding boxes, GradCAM within epochs to check intermediate results and so on.
For more details, please check the official documentation.
# Initializes the W&B run with a config file and W&B run settings.
def wandb_init(fold):
config = {k: v for k, v in dict(vars(CFG)).items() if '__' not in k} # Create config dictionary
config.update({"fold": int(fold)}) # Add fold to config
run = wandb.init(project="llm-science-exam-public",
name=f"fold-{fold}|max_seq-{CFG.sequence_length}|model-{CFG.preset}",
config=config,
group=CFG.comment,
save_code=True)
return run
# Log best result for error analysis
def log_wandb():
wandb.log({'best_acc': best_acc, 'best_acc@3': best_acc3,
'best_loss': best_loss, 'best_epoch': best_epoch})
# Fetch W&B callbacks
def get_wb_callbacks(fold):
# wb_ckpt = wandb.keras.WandbModelCheckpoint(f'fold{fold}.h5',
# monitor='val_accuracy',
# save_best_only=True,
# save_weights_only=False,
# mode='max')
wb_metr = wandb.keras.WandbMetricsLogger()
return [wb_metr] # Return WandB callbacks
⚓ | LR Schedule¶
Implementing a learning rate scheduler is crucial for transfer learning. The learning rate initiates at lr_start and gradually tapers down to lr_min using various techniques, including:
step: Lowering the learning rate in step-wise manner resembling stairs.cos: Utilizing a cosine curve to gradually reduce the learning rate.exp: Exponentially decreasing the learning rate.
Importance: A well-structured learning rate schedule is essential for efficient model training, ensuring optimal convergence and avoiding issues such as overshooting or stagnation.
import math
def get_lr_callback(batch_size=8, mode='cos', epochs=10, plot=False):
lr_start, lr_max, lr_min = 1.0e-6, 0.6e-6 * batch_size, 1e-6
lr_ramp_ep, lr_sus_ep, lr_decay = 2, 0, 0.8
def lrfn(epoch): # Learning rate update function
if epoch < lr_ramp_ep: lr = (lr_max - lr_start) / lr_ramp_ep * epoch + lr_start
elif epoch < lr_ramp_ep + lr_sus_ep: lr = lr_max
elif mode == 'exp': lr = (lr_max - lr_min) * lr_decay**(epoch - lr_ramp_ep - lr_sus_ep) + lr_min
elif mode == 'step': lr = lr_max * lr_decay**((epoch - lr_ramp_ep - lr_sus_ep) // 2)
elif mode == 'cos':
decay_total_epochs, decay_epoch_index = epochs - lr_ramp_ep - lr_sus_ep + 3, epoch - lr_ramp_ep - lr_sus_ep
phase = math.pi * decay_epoch_index / decay_total_epochs
lr = (lr_max - lr_min) * 0.5 * (1 + math.cos(phase)) + lr_min
return lr
if plot: # Plot lr curve if plot is True
plt.figure(figsize=(10, 5))
plt.plot(np.arange(epochs), [lrfn(epoch) for epoch in np.arange(epochs)], marker='o')
plt.xlabel('epoch'); plt.ylabel('lr')
plt.title('LR Scheduler')
plt.show()
return keras.callbacks.LearningRateScheduler(lrfn, verbose=False) # Create lr callback
_=get_lr_callback(CFG.batch_size*CFG.replicas, plot=True)
☎️ | Callbacks¶
The function below will gather all the training callbacks, such as lr_scheduler, model_checkpoint, wandb_logger, and etc.
def get_callbacks(fold):
callbacks = []
lr_cb = get_lr_callback(CFG.batch_size*CFG.replicas) # Get lr callback
ckpt_cb = keras.callbacks.ModelCheckpoint(f'fold{fold}.keras',
monitor='val_accuracy',
save_best_only=True,
save_weights_only=False,
mode='max') # Get Model checkpoint callback
callbacks.extend([lr_cb, ckpt_cb]) # Add lr and checkpoint callbacks
if CFG.wandb: # If WandB is enabled
wb_cbs = get_wb_callbacks(fold) # Get WandB callbacks
callbacks.extend(wb_cbs)
return callbacks # Return the list of callbacks
🤖 | Modeling¶
KerasNLP Classifier¶

The KerasNLP library provides comprehensive, ready-to-use implementations of popular NLP model architectures. It features a variety of pre-trained models including Bert, Roberta, DebertaV3, and more. In this notebook, we'll showcase the usage of DistillBert. However, feel free to explore all available models in the KerasNLP documentation. Also for a deeper understanding of KerasNLP, refer to the informative getting started guide.
Our approach involves using keras_nlp.models.XXClassifier to process each question and option pari (e.g. (Q+A), (Q+B), etc.), generating logits. These logits are then combined and passed through a softmax function to produce the final output.
Classifier for Multiple-Choice Tasks¶
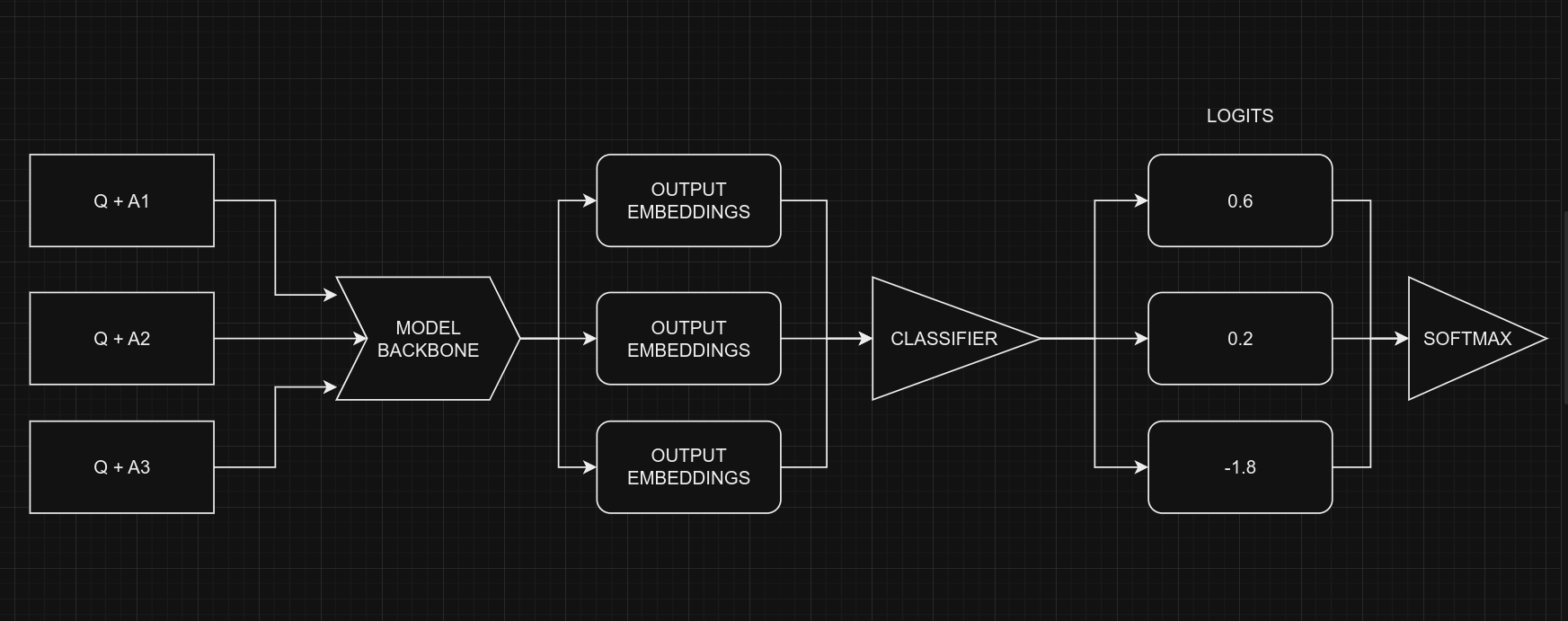
When dealing with multiple-choice questions, instead of giving the model the question and all options together (Q + A + B + C ...), we provide the model with one option at a time along with the question. For instance, (Q + A), (Q + B), and so on. Once we have the prediction scores (logits) for all options, we combine them using the Softmax function to get the ultimate result. If we had given all options at once to the model, the text's length would increase, making it harder for the model to handle. The picture below illustrates this idea:
From a coding perspective, remember that we use the same model for all five options, with shared weights. Despite the figure suggesting five separate models, they are, in fact, one model with shared weights. Another point to consider is the the input shapes of Classifier and MultipleChoice.
- Input shape for Multiple Choice: $(batch\_size, num\_choices, seq\_length)$
- Input shape for Classifier: $(batch\_size, seq\_length)$
Certainly, it's clear that we can't directly give the data for the multiple-choice task to the model because the input shapes don't match. To handle this, we'll use slicing. This means we'll separate the features of each option, like $feature_{(Q + A)}$ and $feature_{(Q + B)}$, and give them one by one to the NLP classifier. After we get the prediction scores $logits_{(Q + A)}$ and $logits_{(Q + B)}$ for all the options, we'll use the Softmax function, like $\operatorname{Softmax}([logits_{(Q + A)}, logits_{(Q + B)}])$, to combine them. This final step helps us make the ultimate decision or choice.
Note that in the classifier, we set
num_classes=1instead of5. This is because the classifier produces a single output for each option. When dealing with five options, these individual outputs are joined together and then processed through a softmax function to generate the final result, which has a dimension of5.
# Selects one option from five
class SelectOption(keras.layers.Layer):
def __init__(self, index, **kwargs):
super().__init__(**kwargs)
self.index = index
def call(self, inputs):
# Selects a specific slice from the inputs tensor
return inputs[:, self.index, :]
def get_config(self):
# For serialize the model
base_config = super().get_config()
config = {
"index": self.index,
}
return {**base_config, **config}
def build_model():
# Define input layers
inputs = {
"token_ids": keras.Input(shape=(5, None), dtype=tf.int32, name="token_ids"),
"padding_mask": keras.Input(shape=(5, None), dtype=tf.int32, name="padding_mask"),
}
# Create a DebertaV3Classifier model
classifier = keras_nlp.models.DebertaV3Classifier.from_preset(
CFG.preset,
preprocessor=None,
num_classes=1 # one output per one option, for five options total 5 outputs
)
logits = []
# Loop through each option (Q+A), (Q+B) etc and compute associted logits
for option_idx in range(5):
option = {k: SelectOption(option_idx, name=f"{k}_{option_idx}")(v) for k, v in inputs.items()}
logit = classifier(option)
logits.append(logit)
# Compute final output
logits = keras.layers.Concatenate(axis=-1)(logits)
outputs = keras.layers.Softmax(axis=-1)(logits)
model = keras.Model(inputs, outputs)
# Compile the model with optimizer, loss, and metrics
model.compile(
optimizer=keras.optimizers.AdamW(5e-6),
loss=keras.losses.CategoricalCrossentropy(label_smoothing=0.02),
metrics=[
keras.metrics.CategoricalAccuracy(name="accuracy"),
keras.metrics.TopKCategoricalAccuracy(k=3, name="accuracy@3")
],
jit_compile=True
)
return model
# with strategy.scope
model = build_model()
Downloading data from https://storage.googleapis.com/keras-nlp/models/deberta_v3_base_en/v1/model.h5 735557816/735557816 ━━━━━━━━━━━━━━━━━━━━ 27s 0us/step
Model Summary¶
model.summary()
Model: "functional_1"
┏━━━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━┳━━━━━━━━━┳━━━━━━━━━━━━━━━━━━━━━━┓ ┃ Layer (type) ┃ Output Shape ┃ Param # ┃ Connected to ┃ ┡━━━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━╇━━━━━━━━━╇━━━━━━━━━━━━━━━━━━━━━━┩ │ padding_mask │ (None, 5, None) │ 0 │ - │ │ (InputLayer) │ │ │ │ ├─────────────────────┼───────────────────┼─────────┼──────────────────────┤ │ token_ids │ (None, 5, None) │ 0 │ - │ │ (InputLayer) │ │ │ │ ├─────────────────────┼───────────────────┼─────────┼──────────────────────┤ │ padding_mask_0 │ (None, None) │ 0 │ padding_mask[0][0] │ │ (SelectOption) │ │ │ │ ├─────────────────────┼───────────────────┼─────────┼──────────────────────┤ │ token_ids_0 │ (None, None) │ 0 │ token_ids[0][0] │ │ (SelectOption) │ │ │ │ ├─────────────────────┼───────────────────┼─────────┼──────────────────────┤ │ padding_mask_1 │ (None, None) │ 0 │ padding_mask[0][0] │ │ (SelectOption) │ │ │ │ ├─────────────────────┼───────────────────┼─────────┼──────────────────────┤ │ token_ids_1 │ (None, None) │ 0 │ token_ids[0][0] │ │ (SelectOption) │ │ │ │ ├─────────────────────┼───────────────────┼─────────┼──────────────────────┤ │ padding_mask_2 │ (None, None) │ 0 │ padding_mask[0][0] │ │ (SelectOption) │ │ │ │ ├─────────────────────┼───────────────────┼─────────┼──────────────────────┤ │ token_ids_2 │ (None, None) │ 0 │ token_ids[0][0] │ │ (SelectOption) │ │ │ │ ├─────────────────────┼───────────────────┼─────────┼──────────────────────┤ │ padding_mask_3 │ (None, None) │ 0 │ padding_mask[0][0] │ │ (SelectOption) │ │ │ │ ├─────────────────────┼───────────────────┼─────────┼──────────────────────┤ │ token_ids_3 │ (None, None) │ 0 │ token_ids[0][0] │ │ (SelectOption) │ │ │ │ ├─────────────────────┼───────────────────┼─────────┼──────────────────────┤ │ padding_mask_4 │ (None, None) │ 0 │ padding_mask[0][0] │ │ (SelectOption) │ │ │ │ ├─────────────────────┼───────────────────┼─────────┼──────────────────────┤ │ token_ids_4 │ (None, None) │ 0 │ token_ids[0][0] │ │ (SelectOption) │ │ │ │ ├─────────────────────┼───────────────────┼─────────┼──────────────────────┤ │ deberta_v3_classif… │ (None, 1) │ 184,42… │ padding_mask_0[0][0… │ │ (DebertaV3Classifi… │ │ │ token_ids_0[0][0], │ │ │ │ │ padding_mask_1[0][0… │ │ │ │ │ token_ids_1[0][0], │ │ │ │ │ padding_mask_2[0][0… │ │ │ │ │ token_ids_2[0][0], │ │ │ │ │ padding_mask_3[0][0… │ │ │ │ │ token_ids_3[0][0], │ │ │ │ │ padding_mask_4[0][0… │ │ │ │ │ token_ids_4[0][0] │ ├─────────────────────┼───────────────────┼─────────┼──────────────────────┤ │ concatenate │ (None, 5) │ 0 │ deberta_v3_classifi… │ │ (Concatenate) │ │ │ deberta_v3_classifi… │ │ │ │ │ deberta_v3_classifi… │ │ │ │ │ deberta_v3_classifi… │ │ │ │ │ deberta_v3_classifi… │ ├─────────────────────┼───────────────────┼─────────┼──────────────────────┤ │ softmax (Softmax) │ (None, 5) │ 0 │ concatenate[0][0] │ └─────────────────────┴───────────────────┴─────────┴──────────────────────┘
Total params: 184,422,913 (5.50 GB)
Trainable params: 184,422,913 (5.50 GB)
Non-trainable params: 0 (0.00 B)
Model Plot¶
keras.utils.plot_model(model, show_shapes=True)
🚂 | Training¶
for fold in CFG.selected_folds:
# Initialize Weights and Biases if enabled
if CFG.wandb:
run = wandb_init(fold)
# Get train and validation datasets
(train_ds, train_df), (valid_ds, valid_df) = get_datasets(fold)
# Get callback functions for training
callbacks = get_callbacks(fold)
# Print training information
print('#' * 50)
print(f'\tFold: {fold + 1} | Model: {CFG.preset}\n\tBatch Size: {CFG.batch_size * CFG.replicas} | Scheduler: {CFG.scheduler}')
print(f'\tNum Train: {len(train_df)} | Num Valid: {len(valid_df)}')
print('#' * 50)
# Clear TensorFlow session and build the model within the strategy scope
K.clear_session()
with strategy.scope():
model = build_model()
# Start training the model
history = model.fit(
train_ds,
epochs=CFG.epochs,
validation_data=valid_ds,
callbacks=callbacks,
steps_per_epoch=int(len(train_df) / CFG.batch_size / CFG.replicas),
)
# Find the epoch with the best validation accuracy
best_epoch = np.argmax(history.history['val_accuracy'])
best_loss = history.history['val_loss'][best_epoch]
best_acc = history.history['val_accuracy'][best_epoch]
best_acc3 = history.history['val_accuracy@3'][best_epoch]
# Print and display best results
print(f'\n{"=" * 17} FOLD {fold} RESULTS {"=" * 17}')
print(f'>>>> BEST Loss : {best_loss:.3f}\n>>>> BEST Acc : {best_acc:.3f}\n>>>> Best Acc@3 : {best_acc3:.3f}\n>>>> BEST Epoch : {best_epoch}')
print('=' * 50)
# Log best result on Weights and Biases (wandb) if enabled
if CFG.wandb:
log_wandb() # Log results
wandb.run.finish() # Finish the run
# display(ipd.IFrame(run.url, width=1080, height=720)) # show wandb dashboard
print("\n\n")
################################################## Fold: 4 | Model: deberta_v3_base_en Batch Size: 2 | Scheduler: cosine Num Train: 660 | Num Valid: 40 ##################################################
/opt/conda/lib/python3.10/site-packages/keras_core/src/backend/jax/numpy.py:103: UserWarning: Explicitly requested dtype int64 requested in arange is not available, and will be truncated to dtype int32. To enable more dtypes, set the jax_enable_x64 configuration option or the JAX_ENABLE_X64 shell environment variable. See https://github.com/google/jax#current-gotchas for more. return jnp.arange(start, stop, step=step, dtype=dtype) /opt/conda/lib/python3.10/site-packages/keras_core/src/backend/jax/core.py:40: UserWarning: Explicitly requested dtype int64 requested in array is not available, and will be truncated to dtype int32. To enable more dtypes, set the jax_enable_x64 configuration option or the JAX_ENABLE_X64 shell environment variable. See https://github.com/google/jax#current-gotchas for more. return jnp.array(x, dtype=dtype) /opt/conda/lib/python3.10/site-packages/keras_core/src/backend/jax/core.py:40: UserWarning: Explicitly requested dtype float64 requested in array is not available, and will be truncated to dtype float32. To enable more dtypes, set the jax_enable_x64 configuration option or the JAX_ENABLE_X64 shell environment variable. See https://github.com/google/jax#current-gotchas for more. return jnp.array(x, dtype=dtype)
Epoch 1/10
/opt/conda/lib/python3.10/site-packages/keras_core/src/backend/jax/numpy.py:103: UserWarning: Explicitly requested dtype int64 requested in arange is not available, and will be truncated to dtype int32. To enable more dtypes, set the jax_enable_x64 configuration option or the JAX_ENABLE_X64 shell environment variable. See https://github.com/google/jax#current-gotchas for more. return jnp.arange(start, stop, step=step, dtype=dtype) /opt/conda/lib/python3.10/site-packages/keras_core/src/backend/jax/core.py:40: UserWarning: Explicitly requested dtype int64 requested in array is not available, and will be truncated to dtype int32. To enable more dtypes, set the jax_enable_x64 configuration option or the JAX_ENABLE_X64 shell environment variable. See https://github.com/google/jax#current-gotchas for more. return jnp.array(x, dtype=dtype)
330/330 ━━━━━━━━━━━━━━━━━━━━ 333s 526ms/step - accuracy: 0.1994 - accuracy@3: 0.6224 - loss: 1.6158 - val_accuracy: 0.3500 - val_accuracy@3: 0.7000 - val_loss: 1.6076 - learning_rate: 1.0000e-06 Epoch 2/10 330/330 ━━━━━━━━━━━━━━━━━━━━ 140s 424ms/step - accuracy: 0.1984 - accuracy@3: 0.6072 - loss: 1.6103 - val_accuracy: 0.4500 - val_accuracy@3: 0.7000 - val_loss: 1.6063 - learning_rate: 1.1000e-06 Epoch 3/10 330/330 ━━━━━━━━━━━━━━━━━━━━ 141s 427ms/step - accuracy: 0.2034 - accuracy@3: 0.6038 - loss: 1.6108 - val_accuracy: 0.4750 - val_accuracy@3: 0.7750 - val_loss: 1.6038 - learning_rate: 1.2000e-06 Epoch 4/10 330/330 ━━━━━━━━━━━━━━━━━━━━ 137s 416ms/step - accuracy: 0.2413 - accuracy@3: 0.6476 - loss: 1.6009 - val_accuracy: 0.5000 - val_accuracy@3: 0.8000 - val_loss: 1.5916 - learning_rate: 1.1959e-06 Epoch 5/10 330/330 ━━━━━━━━━━━━━━━━━━━━ 126s 383ms/step - accuracy: 0.2401 - accuracy@3: 0.6739 - loss: 1.5873 - val_accuracy: 0.4750 - val_accuracy@3: 0.8250 - val_loss: 1.5087 - learning_rate: 1.1841e-06 Epoch 6/10 330/330 ━━━━━━━━━━━━━━━━━━━━ 139s 421ms/step - accuracy: 0.3725 - accuracy@3: 0.7924 - loss: 1.4761 - val_accuracy: 0.5250 - val_accuracy@3: 0.8000 - val_loss: 1.3829 - learning_rate: 1.1655e-06 Epoch 7/10 330/330 ━━━━━━━━━━━━━━━━━━━━ 126s 382ms/step - accuracy: 0.4781 - accuracy@3: 0.8305 - loss: 1.3143 - val_accuracy: 0.5000 - val_accuracy@3: 0.8250 - val_loss: 1.2930 - learning_rate: 1.1415e-06 Epoch 8/10 330/330 ━━━━━━━━━━━━━━━━━━━━ 126s 381ms/step - accuracy: 0.5179 - accuracy@3: 0.8737 - loss: 1.2052 - val_accuracy: 0.5250 - val_accuracy@3: 0.8250 - val_loss: 1.2626 - learning_rate: 1.1142e-06 Epoch 9/10 330/330 ━━━━━━━━━━━━━━━━━━━━ 126s 381ms/step - accuracy: 0.5370 - accuracy@3: 0.8802 - loss: 1.1658 - val_accuracy: 0.5000 - val_accuracy@3: 0.8750 - val_loss: 1.2707 - learning_rate: 1.0858e-06 Epoch 10/10 330/330 ━━━━━━━━━━━━━━━━━━━━ 126s 381ms/step - accuracy: 0.5722 - accuracy@3: 0.9001 - loss: 1.0415 - val_accuracy: 0.4750 - val_accuracy@3: 0.8500 - val_loss: 1.2567 - learning_rate: 1.0585e-06 ================= FOLD 3 RESULTS ================= >>>> BEST Loss : 1.383 >>>> BEST Acc : 0.525 >>>> Best Acc@3 : 0.800 >>>> BEST Epoch : 5 ================================================== ################################################## Fold: 5 | Model: deberta_v3_base_en Batch Size: 2 | Scheduler: cosine Num Train: 660 | Num Valid: 40 ##################################################
/opt/conda/lib/python3.10/site-packages/keras_core/src/backend/jax/numpy.py:103: UserWarning: Explicitly requested dtype int64 requested in arange is not available, and will be truncated to dtype int32. To enable more dtypes, set the jax_enable_x64 configuration option or the JAX_ENABLE_X64 shell environment variable. See https://github.com/google/jax#current-gotchas for more. return jnp.arange(start, stop, step=step, dtype=dtype) /opt/conda/lib/python3.10/site-packages/keras_core/src/backend/jax/core.py:40: UserWarning: Explicitly requested dtype int64 requested in array is not available, and will be truncated to dtype int32. To enable more dtypes, set the jax_enable_x64 configuration option or the JAX_ENABLE_X64 shell environment variable. See https://github.com/google/jax#current-gotchas for more. return jnp.array(x, dtype=dtype) /opt/conda/lib/python3.10/site-packages/keras_core/src/backend/jax/core.py:40: UserWarning: Explicitly requested dtype float64 requested in array is not available, and will be truncated to dtype float32. To enable more dtypes, set the jax_enable_x64 configuration option or the JAX_ENABLE_X64 shell environment variable. See https://github.com/google/jax#current-gotchas for more. return jnp.array(x, dtype=dtype)
Epoch 1/10 330/330 ━━━━━━━━━━━━━━━━━━━━ 323s 518ms/step - accuracy: 0.1652 - accuracy@3: 0.6172 - loss: 1.6123 - val_accuracy: 0.2000 - val_accuracy@3: 0.5250 - val_loss: 1.6096 - learning_rate: 1.0000e-06 Epoch 2/10 330/330 ━━━━━━━━━━━━━━━━━━━━ 141s 428ms/step - accuracy: 0.2285 - accuracy@3: 0.6456 - loss: 1.6089 - val_accuracy: 0.2500 - val_accuracy@3: 0.5250 - val_loss: 1.6086 - learning_rate: 1.1000e-06 Epoch 3/10 330/330 ━━━━━━━━━━━━━━━━━━━━ 141s 428ms/step - accuracy: 0.1724 - accuracy@3: 0.5866 - loss: 1.6126 - val_accuracy: 0.3000 - val_accuracy@3: 0.7500 - val_loss: 1.6068 - learning_rate: 1.2000e-06 Epoch 4/10 330/330 ━━━━━━━━━━━━━━━━━━━━ 126s 382ms/step - accuracy: 0.2106 - accuracy@3: 0.6235 - loss: 1.6059 - val_accuracy: 0.2250 - val_accuracy@3: 0.7750 - val_loss: 1.5980 - learning_rate: 1.1959e-06 Epoch 5/10 330/330 ━━━━━━━━━━━━━━━━━━━━ 141s 426ms/step - accuracy: 0.2491 - accuracy@3: 0.6980 - loss: 1.5886 - val_accuracy: 0.3250 - val_accuracy@3: 0.8250 - val_loss: 1.5635 - learning_rate: 1.1841e-06 Epoch 6/10 330/330 ━━━━━━━━━━━━━━━━━━━━ 141s 426ms/step - accuracy: 0.3796 - accuracy@3: 0.7816 - loss: 1.5175 - val_accuracy: 0.3750 - val_accuracy@3: 0.8500 - val_loss: 1.4857 - learning_rate: 1.1655e-06 Epoch 7/10 330/330 ━━━━━━━━━━━━━━━━━━━━ 126s 383ms/step - accuracy: 0.4068 - accuracy@3: 0.7964 - loss: 1.3951 - val_accuracy: 0.3250 - val_accuracy@3: 0.8500 - val_loss: 1.4397 - learning_rate: 1.1415e-06 Epoch 8/10 330/330 ━━━━━━━━━━━━━━━━━━━━ 126s 383ms/step - accuracy: 0.4998 - accuracy@3: 0.8215 - loss: 1.2799 - val_accuracy: 0.3000 - val_accuracy@3: 0.8500 - val_loss: 1.4464 - learning_rate: 1.1142e-06 Epoch 9/10 330/330 ━━━━━━━━━━━━━━━━━━━━ 127s 383ms/step - accuracy: 0.4961 - accuracy@3: 0.8534 - loss: 1.2225 - val_accuracy: 0.3250 - val_accuracy@3: 0.8500 - val_loss: 1.4102 - learning_rate: 1.0858e-06 Epoch 10/10 330/330 ━━━━━━━━━━━━━━━━━━━━ 126s 382ms/step - accuracy: 0.5511 - accuracy@3: 0.9054 - loss: 1.1470 - val_accuracy: 0.3500 - val_accuracy@3: 0.9000 - val_loss: 1.4517 - learning_rate: 1.0585e-06 ================= FOLD 4 RESULTS ================= >>>> BEST Loss : 1.486 >>>> BEST Acc : 0.375 >>>> Best Acc@3 : 0.850 >>>> BEST Epoch : 5 ==================================================
🧪 | Prediction¶
# Make predictions using the trained model on last validation data
predictions = model.predict(
valid_ds,
batch_size=min(CFG.batch_size * CFG.replicas * 2, len(valid_df)), # max batch size = valid size
verbose=1
)
20/20 ━━━━━━━━━━━━━━━━━━━━ 33s 86ms/step
# Format predictions and true answers
pred_answers = np.array(list('ABCDE'))[np.argsort(-predictions)][:, 0]
true_answers = valid_df.answer.values
# Check 5 Predictions
print("# Predictions\n")
for i in range(5):
row = valid_df.iloc[i]
question = row.prompt
pred_answer = pred_answers[i]
true_answer = true_answers[i]
print(f"❓ Question {i+1}:\n{question}\n")
print(f"✅ True Answer: {true_answer}\n >> {row[true_answer]}\n")
print(f"🤖 Predicted Answer: {pred_answer}\n >> {row[pred_answer]}\n")
print("-"*90, "\n")
# Predictions ❓ Question 1: Which of the following statements accurately describes the impact of Modified Newtonian Dynamics (MOND) on the observed "missing baryonic mass" discrepancy in galaxy clusters? ✅ True Answer: D >> MOND is a theory that reduces the discrepancy between the observed missing baryonic mass in galaxy clusters and the measured velocity dispersions from a factor of around 10 to a factor of about 2. 🤖 Predicted Answer: B >> MOND is a theory that increases the discrepancy between the observed missing baryonic mass in galaxy clusters and the measured velocity dispersions from a factor of around 10 to a factor of about 20. ------------------------------------------------------------------------------------------ ❓ Question 2: Which of the following statements accurately describes the relationship between the dimensions of a diffracting object and the angular spacing of features in the diffraction pattern? ✅ True Answer: D >> The angular spacing of features in the diffraction pattern is inversely proportional to the dimensions of the object causing the diffraction. Therefore, if the diffracting object is smaller, the resulting diffraction pattern will be wider. 🤖 Predicted Answer: E >> The angular spacing of features in the diffraction pattern is directly proportional to the square root of the dimensions of the object causing the diffraction. Therefore, if the diffracting object is smaller, the resulting diffraction pattern will be slightly narrower. ------------------------------------------------------------------------------------------ ❓ Question 3: Which of the following statements accurately describes the dimension of an object in a CW complex? ✅ True Answer: A >> The dimension of an object in a CW complex is the largest n for which the n-skeleton is nontrivial, where the empty set is considered to have dimension -1 and the boundary of a discrete set of points is the empty set. 🤖 Predicted Answer: C >> The dimension of an object in a CW complex is the smallest n for which the n-skeleton is nontrivial. The empty set is given a dimension of -1, while the boundary of a discrete set of points is assigned a dimension of 0. ------------------------------------------------------------------------------------------ ❓ Question 4: What is the term used in astrophysics to describe light-matter interactions resulting in energy shifts in the radiation field? ✅ True Answer: C >> Reddening 🤖 Predicted Answer: B >> Redshifting ------------------------------------------------------------------------------------------ ❓ Question 5: What is Martin Heidegger's view on the relationship between time and human existence? ✅ True Answer: B >> Martin Heidegger believes that humans do not exist inside time, but that they are time. The relationship to the past is a present awareness of having been, and the relationship to the future involves anticipating a potential possibility, task, or engagement. 🤖 Predicted Answer: A >> Martin Heidegger believes that humans exist within a time continuum that is infinite and does not have a defined beginning or end. The relationship to the past involves acknowledging it as a historical era, and the relationship to the future involves creating a world that will endure beyond one's own time. ------------------------------------------------------------------------------------------
✍️ | Reference¶
- Multiple Choice with HF @johnowhitaker
- Keras NLP
- BirdCLEF23: Pretraining is All you Need [Train] by @awsaf49
- Triple Stratified KFold with TFRecords by @cdeotte
!rm -r /kaggle/working/wandb